What is DeNovoDB?
DeNovoDB: A Comprehensive Resource for Investigating De Novo Emerged Genes
DeNovoDB is a centralized web-based resource designed to facilitate the identification, and exploration of de novo emerged genes (i.e. from noncoding DNA) using the DENSE (DE Novo emerged genes SEarch) pipeline. It provides two primary functionalities:
1. Exploring Pre-computed DENSE Results:
DeNovoDB houses pre-computed DENSE data for several model organisms. This comprehensive dataset encompasses the classification of CDSs according to various DENSE strategies. The data is readily available for filtering, and downloading, empowering researchers to tailor their analyses to specific research questions.
2. Interactive DENSE Web Server:
DeNovoDB goes beyond pre-computed data by offering an interactive web server that allows users to run DENSE directly within the platform, bypassing the phylostratigraphy step. This streamlined approach simplifies the analysis process and eliminates the need for external software installation. Users simply provide a list of taxonomically restricted genes (see more), and the web server automatically initiates the DENSE analysis. For model organisms within DeNovoDB, users can easily obtain TRGs lists using the various filters of the database.
A Resource for Exploring DEG Diversification and Enhancing Collaboration
DeNovoDB empowers researchers to explore de novo genes across diverse organisms. Its centralized platform facilitates the identification of patterns and variations in DEG emergence, aiding in understanding the mechanisms driving de novo gene emergence and their contribution to biological diversity. Additionally, DeNovoDB promotes reproducibility and collaboration by providing transparent and accessible data and analysis workflows.
DENSE concepts
TRG
DENSE employs phylostratygraphy to identify taxonomically restricted genes (TRGs), which are genes that are unique to a small taxonomic group. It uses GenEra (Barrera-Redondo et al., 2023), a phylostratygraphy tool, to assess the evolutionary history of genes. By tracing the ancestry of gene homologs across different species, phylostratygraphy assigns a theoretical age to each gene. A gene is considered TRG if its most recent common ancestor (MRCA) lies within a specific taxonomic group, typically the genus (default parameter). However, this threshold can be customized, as demonstrated in DeNovoDB, where TRG thresholds for human and mouse extend beyond the genus level. This flexibility allows researchers to tailor their analysis to specific taxonomic interests.
Outgroups
A genome labeled as "outgroup" is a genome where a given gene is absent and which branches in the tree **after** the last genome where the gene is present.
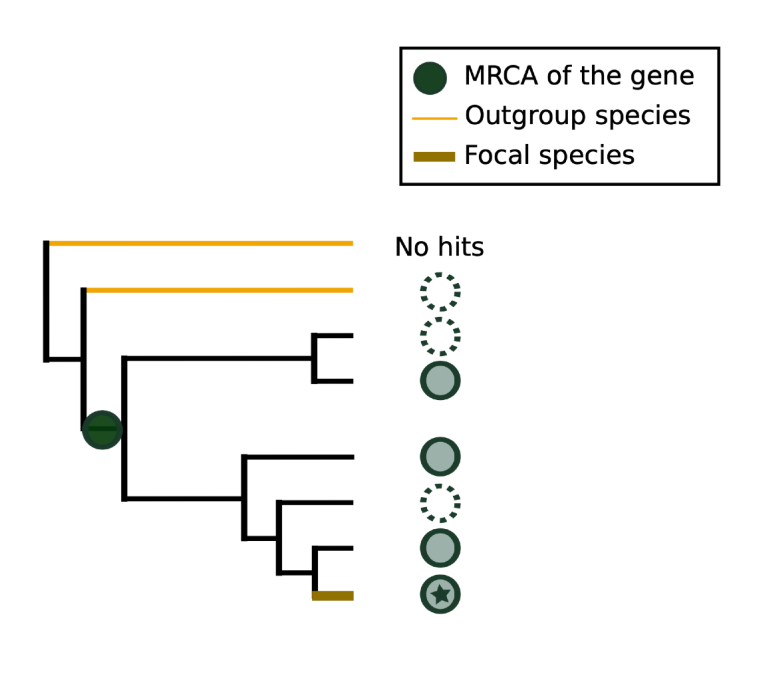
Microsynteny
DENSE employs a microsynteny to validate synteny conservation between a gene and its noncoding homologs, addressing limitations posed by chromosomal rearrangements and fragmented assemblies. It defines two gene windows of the same size, one upstream and one downstream of each gene, and examines whether at least one orthologous gene from each window can be detected in the corresponding target window of an outgroup species. The default window size in 4 genes (4 upstream, 4 downstream), and the default number of required anchor pairs is set to one.
Examples of Synteny Conservation Checking
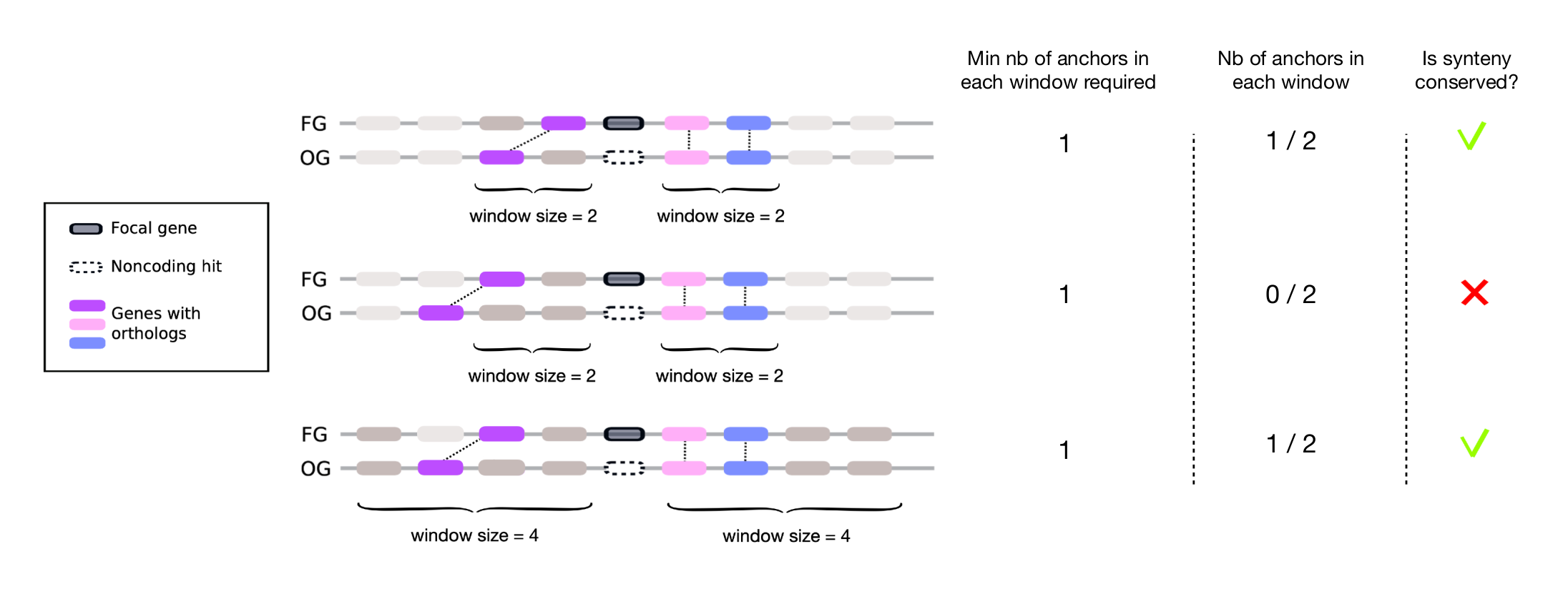
Consider a de novo gene candidate (black) in a focal genome (FG) and its homologous noncoding hit (white, dashed borders). The following examples illustrate the process of synteny conservation checking:
- In the first example, one gene from the upstream focal window (purple orthologous pair forming the upstream anchor) is retrieved in the upstream target window. Similarly, both genes from the downstream focal window (orthologous pairs in pink and blue forming the downstream anchors) are retrieved in the downstream target window. This indicates that synteny is conserved since each window is associated with at least one anchor gene.
- In the second example, no anchor is found within the upstream window (i.e., the purple ortholog is located outside the target window). Consequently, the conservation of synteny is not validated.
- In the last example, the window size is extended to four genes, enabling the purple ortholog in the outgroup species to be retrieved. This results in the presence of an anchor in each window, indicating that synteny is conserved.
Jobs & uploaded data
When you upload your data on this web service to run the DENSE pipeline, your data will be stored on the I2BC's file system for up to 30 days in an area with restricted access. After 30 days, all files related to your job will be deleted and unaccessible to you but might remain recoverable for up to 6 months after job sumbission thanks to the I2BC's file system backup policy.
All your files will be stored within a subfolder of this restricted access area with a unique and random identifier (uuid) corresponding to the id that you are given when submitting your job. You can follow or access your results anytime using the access link with the following format: /denovodb/dense-run/your-uuid/follow (replace 'your-uuid' by your job id).
How to cite?
This work
DENSE: Roginski, P. et al. DE Novo emerged gene SEarch in Eukaryotes with DENSE Genome Biol Evol. 2024 Aug;16(8). doi: 10.1093/gbe/evae159
Link to DENSE GitHub repository: https://github.com/Proginski/dense
Tools used in this work
Nextflow: Di Tommaso, P. et al. Nextflow enables reproducible computational workflows. Nat Biotechnol. 2017 Apr 11;35(4):316-319. doi: 10.1038/nbt.3820.
AWK: Aho, A. V. et al. (1979). Awk — a pattern scanning and processing language. Software: Practice and Experience, 9(4), 267-279.
BEDTools: Aaron R. Quinlan, Ira M. Hall, BEDTools: a flexible suite of utilities for comparing genomic features, Bioinformatics, Volume 26, Issue 6, March 2010, Pages 841–842.
GffRead: Pertea G and Pertea M., GFF Utilities: GffRead and GffCompare [version 2; peer review: 3 approved]. F1000Research 2020, 9:304.
GenEra: Barrera-Redondo, J. et al. (2023). Uncovering gene-family founder events during major evolutionary transitions in animals, plants and fungi using GenEra. Genome Biology, 24(1), 54. doi: 10.1186/s13059-023-02895-z.
BLAST: Altschul, S.F. et al., (1990) “Basic local alignment search tool.” J. Mol. Biol. 215:403-410.
MultiQC: Ewels P. et al., MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016 Oct 1;32(19):3047-8. doi: 10.1093/bioinformatics/btw354. Epub 2016 Jun 16. PubMed
Docker: Kurtzer GM, et al., Singularity: Scientific containers for mobility of compute. PLoS One. 2017 May 11;12(5):e0177459. doi: 10.1371/journal.pone.0177459. eCollection 2017. PubMed
Singularity: Merkel, D. (2014). Docker: lightweight linux containers for consistent development and deployment. Linux Journal, 2014(239), 2. doi: 10.5555/2600239.2600241.
Other references
This site was generated using Django (Version 4.1.7) [Computer Software]. (2023). Retrieved from https://www.djangoproject.com/.
The species pictures on the front page were fetched and modified from various sources with Creative Common Licences under the following links: A. thaliana, C. elegans and D. melanogaster from Wikipedia, and H. sapiens, M. musculus, O. stavia and S. cerevisiae from Phylopic.
Contact information
For help or to report a problem, please email us at contact-bioi2@i2bc.paris-saclay.fr.
For issues that are more specific to the DENSE pipeline, you can also generate an Issue on the DENSE GitHub page.