/!\ Please note that services will be unavailable for 3 days (17/11/2025 to 19/11/2025) for technical reasons. Sorry for the inconvenience.
Convert MSA file formats
What it does? converts Multiple Sequence Alignment (MSA) files from a3m format to fasta format using the HH-suite's reformat.pl script.
This is particularly useful with AlphaFold/Colabfold/MMseqs2 a3m outputs as many visualisation tools don't support this format.
Generate MSAs from sequences

What it does? searches for homologs of a given protein sequence within the UniRef30 using MMseqs2.
Run evolutionary rate analysis on proteins
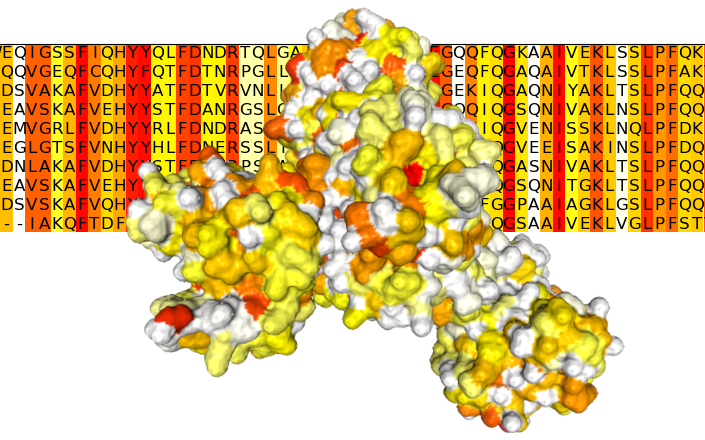
What it does? analyses evolutionary rates of proteins using Rate4Site.
Process outline:
- If not given, MSAs are computed using MMseqs2 on the UniRef30 database using the sequence extracted from the given structure.
- By default, MSAs from different protein chains are split and dealt with independently (this behaviour can be changed with the "coupled" option).
- First, they are filtered for sequence identity redundancy using hhfilter (default threshold is 80%).
- The N most similar sequences (based on sequence identity) with the query sequence are extracted from the filtered MSAs to run Rate4Site (N=100 by default).
- Rate4Site scores are normalised between 0 and 100 and transfered into the occupancy/bfactor field of the given protein structures (pairing between MSA query sequence and protein sequence is done using sequence identity).
Code availability: MSA-tools project on Github
References
Tools used
This site was generated using Django and django modules. Protein structures are displayed in three-dimensions using the WebGL-based NGL Viewer plugin. Many thanks also to the RPBS platform and their NGL Viewer code from which we inspired ourselves to create the protein visualisations in NGL Viewer.
When running jobs, we use MMseqs2 (commit 4148e09, 30/01/2023) to generate multiple sequence alignments using ColabFold's pipeline (v1.5.2 commit 3574273, 24/02/2023) on their UniRef30 (v2202) database, we use the HH-suite (v3.3.0) to filter and reduce the alignments, and we use Rate4Site (v3.0.0) to compute evolutionary rates based on these sequence alignments.
-
Django v4.1.6 [Computer Software]. (2023).
https://www.djangoproject.com/ -
Bootstrap v4.1.3
Github. https://github.com/twbs/bootstrap -
AS Rose, AR Bradley, Y Valasatava, JM Duarte, A Prlić and PW Rose.
Web-based molecular graphics for large complexes.
ACM Proceedings of the 21st International Conference on Web3D Technology, 2016.
doi: 10.1145/2945292.2945324 -
AS Rose and PW Hildebrand.
NGL Viewer: a web application for molecular visualization.
Nucl Acids Res, 2015.
doi: 10.1093/nar/gkv402 -
AS Rose and PW Hildebrand.
NGL Viewer: a web application for molecular visualization.
Nucl Acids Res, 2015.
doi: 10.1093/nar/gkv402 -
Mirdita, M., Schütze, K., Moriwaki, Y. et al. ColabFold: making protein folding accessible to all.
Nat. Methods 19, 679-682 (2022).
doi: https://doi.org/10.1038/s41592-022-01488-1 -
Mirdita, M., Steinegger, M. and Söding, J.. MMseqs2 desktop and local web server app for fast, interactive sequence searches.
Bioinformatics 35(16), 2856-2858 (2019).
doi: https://doi.org/10.1093/bioinformatics/bty1057 -
Mirdita, M., von den Driesch, L., Galiez, C., Martin, M.J., Söding, J., Steinegger, M..
Uniclust databases of clustered and deeply annotated protein sequences and alignments.
Nucleic Acids Res. 45(D1), D170-D176 (2017).
doi: https://doi.org/10.1093/nar/gkw1081 -
Steinegger M, Meier M, Mirdita M, Vöhringer H, Haunsberger S J, and Söding J (2019)
HH-suite3 for fast remote homology detection and deep protein annotation,
BMC Bioinformatics, 473.
doi: https://doi.org/10.1186/s12859-019-3019-7 -
Tal Pupko, Rachel E. Bell, Itay Mayrose, Fabian Glaser, Nir Ben-Tal,
Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues,
Bioinformatics, Volume 18, Issue suppl_1, July 2002, Pages S71–S77
doi: https://doi.org/10.1093/bioinformatics/18.suppl_1.S71
Acknowledgements
We thank Hugo Pointier, an M2 intern at BIOI2, for leading this project to completion. We also thank Martin Pagani from the Informatics support team of the I2BC for his precious help in setting up the MSAviewer plugin. Also a big thanks to Jessica Andreani, Raphaël Guerois and Diego Zea from the Molecular Assemblies and Genome Integrity team of the I2BC for sharing their scripts and for their input in developping this website. Many thanks also to the I2BC and it's IT support team for making the computer cluster and the webserver available.