What is ORENZA?
ORENZA is a relationnal database of ORphan ENZyme Activities. It regroups a list of all referenced enzyme activities and their "orphan status". This web site allows you to easily browse and search through them, by activity, creation date, pathway or class.
What are orphan enzyme activities?
Orphan enzyme activities correpond to activities that were detected experimentally that are linked to no known amino acid sequence listed in the major public databases (i.e. UniProt & PDB). These activities are defined by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB).
How is ORENZA built?
The essence of ORENZA is to bring together and analyse cross-mapping of databases. ORENZA uses and links 5 major databases:
- Reference database of enzyme activities: The up-to-date list of all referenced enzyme activities is fetched from ExplorEnz, in which each activity is labelled with a 4-level classification system from NC-IUBMB: the EC number, in the format EC A.B.C.D where A, B and C correspond to class, sub- and subsub-class and D corresponds to a serial number specific to each activity within that sub-subclass.
- Main sequence databases: We scan through the UniProtKB/Swiss-Prot, UniProtKB/TrEMBL and the Protein Data Bank (PDB) and link each EC number referenced in these databases with the list of EC numbers from ExplorEnz. Any leftover EC numbers (i.e. activities) are then labelled as orphan in ORENZA.
- Brenda & KEGG: We also scan through Brenda and KEGG databases to link mentioned EC numbers with occurences in specific species and metabolic pathways respectively.
Scripts used to update ORENZA can be found on the I2BC's GitHub page.
How to use ORENZA?
Main table

All EC numbers (according to the NC-IUBMB nomenclature) and associated data are listed and can be easily browsed using the "EC number" page. This data entails its common name from ExplorEnz, its "orphan status" (True/False), the number of entries in Swiss-Prot, TrEMBL and PDB that are linked to it, and the number of species in Brenda that are associated with it.
Accessory tables

The "EC classes" page allows yout o browse ORENZA entries hierarchically, starting from ExplorEnz's classes, through sub- and subsub-classes, down to the EC Numbers table containing only activities from within the given class, sub- and subsub-class. The class, sub- and subsub-class tables also provide the number of activities within each group and how many of them are orphan.
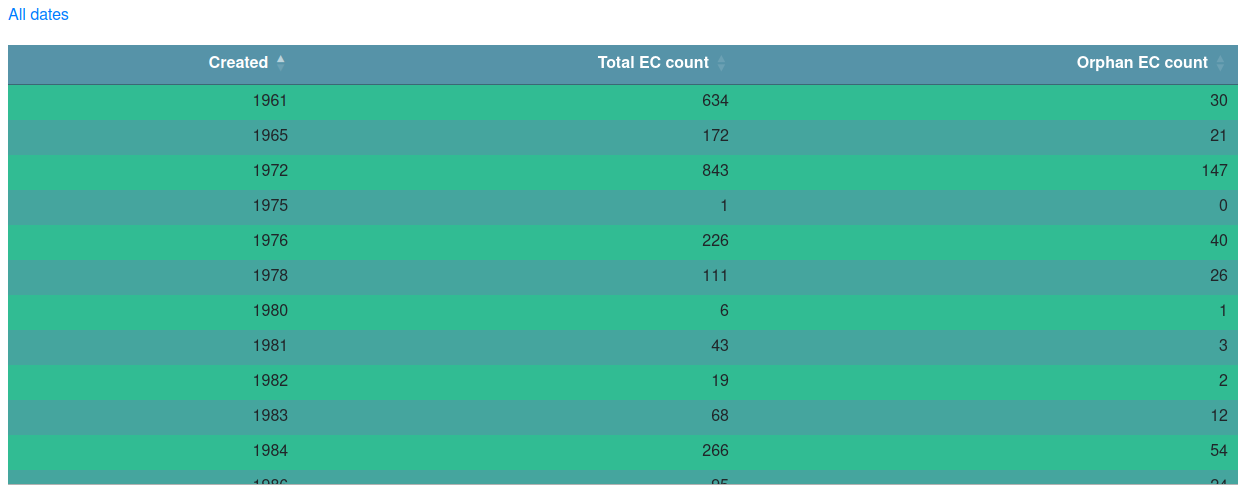
The "Creation date" page allows you to browse ORENZA entries by creation date (i.e. date of entry of an activity in ExplorEnz). Similarly to the "EC class" table, it also provides the count of activities within for each date and the number of orphans within them.
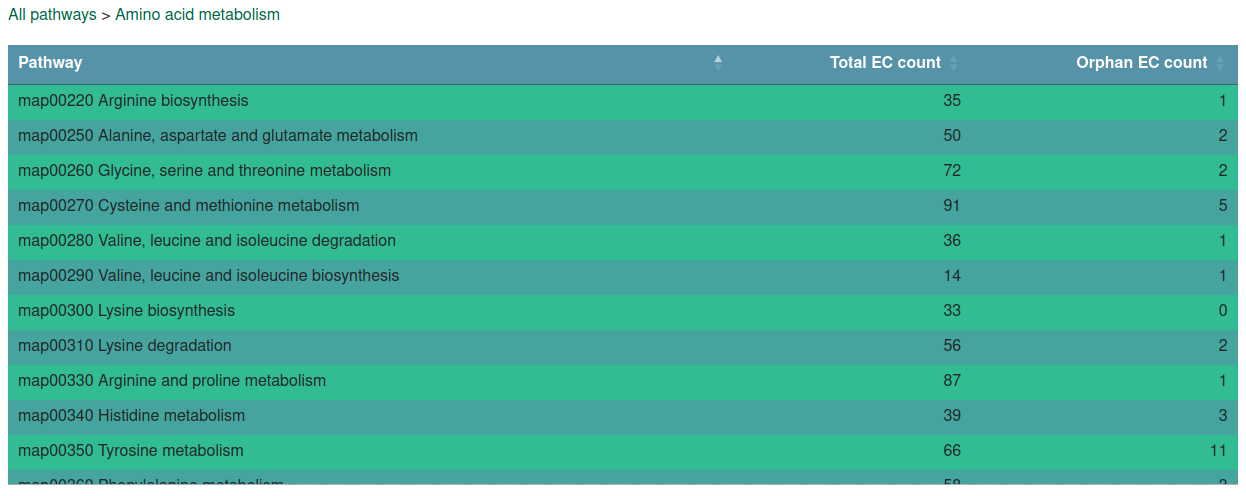
The "Kegg pathways" page allows you to browse ORENZA entries hierarchically, starting from KEGG's pathway classes, through its pathways, down to the EC Numbers table containing only activities from within the given pathway class or pathway. The pathway class and pathway tables also provide the number of activities within each group and how many of them are orphan.
Details on a given activity
Each EC number has a dedicated page listing all its characteristics collected from ExplorEnz and all accession numbers of Swiss-Prot, TrEMBL and PDB in which it appears, as well as the list of species that it was reported in from Brenda.
Searching through ORENZA
All tables and accession lists have an associated search bar at the top right corner in order to filter by EC number, enzyme activity name or species. Columns can also be ordered in ascending/descending order for easier navigation.
Summary plots
The "Summary plots" page can be used to visualise the evolution of the number of referenced enzyme activities and the counts of orphan ones in ExplorEnz over time, as well as orphan/non-orphan proportions by activity creation date.
Version
As of August 2024, ORENZA will be updated every 6 months thanks to an automated pipeline. The current version of ORENZA is 2025-07 ("year-month" of latest update).
This version is based on data from:
- Explorenz: 2025.05
- Brenda: Release 2025.1 (May 2025)
- Kegg: Release 115.0, July 1, 2025
- UniprotKB/Swiss-Prot: Release 2025_03 of 18-Jun-2025
- UniprotKB/TrEMBL: Release 2025_03 of 18-Jun-2025
- PDB: Downloaded on 2025-07
References
ORENZA was originally developed by Olivier Lespinet and Bernard Labedan. Regular update scripts and the current website were developed by Hugo Pointier (M2 intern) and the BIOI2 bioinformatics facility of the I2BC, France.This work:
-
ORENZA: a web resource for studying ORphan ENZyme Activities.
Lespinet O and Labedan B,
BMC Bioinformatics. 2006 Oct 6;7:436.
doi: 10.1186/1471-2105-7-436
-
Orphan enzymes could be an unexplored reservoir of new drug targets.
Lespinet O and Labedan B,
Drug Discov Today. 2006 Apr;11(7-8):300-5.
doi: 10.1016/j.drudis.2006.02.002 -
Puzzling over orphan enzymes.
Lespinet O and Labedan B,
Cell Mol Life Sci. 2006 Mar;63(5):517-23.
doi: 10.1007/s00018-005-5520-6 -
Orphan enzymes?
Lespinet O and Labedan B,
Science. 2005 Jan 7;307(5706):42.
doi: 10.1126/science.307.5706.42a
-
ExplorEnz:
McDonald, A.G., Boyce, S. and Tipton, K.F.
ExplorEnz: the primary source of the IUBMB enzyme list.
Nucleic Acids Res. 37, D593–D597 (2009).
doi: 10.1093/nar/gkn582 -
Brenda:
Chang A., Jeske L., Ulbrich S., Hofmann J., Koblitz J., Schomburg I., Neumann-Schaal M., Jahn D., Schomburg D.
BRENDA, the ELIXIR core data resource in 2021: new developments and updates. (2021), Nucleic Acids Res., 49:D498-D508.
doi: 10.1093/nar/gkaa1025
Distributed under CC-BY license: https://creativecommons.org/licenses/by/4.0/ -
UniProt:
The UniProt Consortium
UniProt: the Universal Protein Knowledgebase in 2023
Nucleic Acids Res. 51:D523–D531 (2023)
doi: 10.1093/nar/gkac1052 -
KEGG:
Kanehisa M, Goto S.
KEGG: kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 2000 Jan 1;28(1):27-30.
doi: 10.1093/nar/28.1.27 -
PDB:
The Protein Data Bank
H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne (2000)
Nucleic Acids Research, 28: 235-242.
doi: 10.1093/nar/28.1.235
This site was generated using Django and django modules. Charts were plotted using Chart.js and ORENZA tables are rendered with dataTables.js.
-
Django v4.1.6 [Computer Software]. (2023).
https://www.djangoproject.com/ -
Bootstrap v4.1.3
Github. https://github.com/twbs/bootstrap -
jQuery v3.7.1
Github. https://github.com/jquery/jquery -
DataTables v2.1.0
http://datatables.net -
Chart.js (2024).
https://www.chartjs.org
Contacts
ORENZA is currently maintained by Olivier Lespinet and the BIOI2 bioinformatics facility of the I2BC. Any comments or suggestions can be directly addressed to either one of them.
Acknowledgements
We thank Hugo Pointier, an M2 intern at BIOI2, for leading the ORENZA update project to completion.