Getting started with Snakemake
Objective 1:
Learn how to tell Snakemake that multiple processors are available locally using the
--cores option.Where to start?
Remember
--cores/-c from your previous commands? This option tells Snakemake how many processors (/CPUs/cores/threads) it may use to run your jobs locally. It’s a mandatory option so if you run Snakemake without it, you’ll get an error message.john.doe@node06:/data/work/I2BC/john.doe/snakemake_tutorial$ qstat -u $USER -wn1
pbsserver:
Req'd Req'd Elap
Job ID Username Queue Jobname SessID NDS TSK Memory Time S Time
------------------------------ --------------- --------------- --------------- -------- ---- ----- ------ ----- - -----
747716.pbsserver john.doe common STDIN 825631 1 1 2gb 02:00 R 00:18 node06/3
john.doe@node06:/data/work/I2BC/john.doe/snakemake_tutorial$ logout qsub: job 747716.pbsserver completed
john.doe@cluster-i2bc:~$ qsub -I -q common -l ncpus=2 qsub: waiting for job 747738.pbsserver to start qsub: job 747738.pbsserver ready john.doe@node06:~$ qstat -u $USER -wn1 pbsserver: Req'd Req'd Elap Job ID Username Queue Jobname SessID NDS TSK Memory Time S Time ------------------------------ --------------- --------------- --------------- -------- ---- ----- ------ ----- - ----- 747738.pbsserver john.doe common STDIN 825631 1 2 2gb 02:00 R 00:18 node06/3
john.doe@node06:~$ cd /data/work/I2BC/$USER/snakemake_tutorial john.doe@node06:/data/work/I2BC/john.doe/snakemake_tutorial$ module load snakemake/snakemake-8.4.6 fastqc/fastqc_v0.11.5 nodes/multiqc-1.9
Run Snakemake on 2 local processors
Running Snakemake on 2 processors instead of just one is quite straightforward (we’ll add
-R fastqc to force Snakemake to re-run everything so you can observe the changes):
snakemake -s ex1b_o3.smk -p --configfile ex1.yml -R fastqc --cores 2
Your output should look like this:
Assuming unrestricted shared filesystem usage for local execution.
Building DAG of jobs...
Using shell: /usr/bin/bash
Provided cores: 2
Rules claiming more threads will be scaled down.
Job stats:
job count
------- -------
all 1
fastqc 6
multiqc 1
total 8
Select jobs to execute...
Execute 2 jobs...
[Wed Feb 21 13:37:38 2024]
localrule fastqc:
input: Data/SRR3105698_chr18.fastq.gz
output: FastQC/SRR3105698_chr18_fastqc.zip, FastQC/SRR3105698_chr18_fastqc.html
log: Logs/SRR3105698_chr18_fastqc.std, Logs/SRR3105698_chr18_fastqc.err
jobid: 2
reason: Forced execution
wildcards: sample=SRR3105698_chr18
resources: tmpdir=/var/tmp/pbs.747714.pbsserver
[Wed Feb 21 13:37:38 2024]
localrule fastqc:
input: Data/SRR3105699_chr18.fastq.gz
output: FastQC/SRR3105699_chr18_fastqc.zip, FastQC/SRR3105699_chr18_fastqc.html
log: Logs/SRR3105699_chr18_fastqc.std, Logs/SRR3105699_chr18_fastqc.err
jobid: 6
reason: Forced execution
wildcards: sample=SRR3105699_chr18
resources: tmpdir=/var/tmp/pbs.747714.pbsserver
[Wed Feb 21 13:37:52 2024]
Finished job 6.
1 of 8 steps (12%) done
Select jobs to execute...
Execute 1 jobs...
[Wed Feb 21 13:37:52 2024]
localrule fastqc:
input: Data/SRR3099586_chr18.fastq.gz
output: FastQC/SRR3099586_chr18_fastqc.zip, FastQC/SRR3099586_chr18_fastqc.html
log: Logs/SRR3099586_chr18_fastqc.std, Logs/SRR3099586_chr18_fastqc.err
jobid: 1
reason: Forced execution
wildcards: sample=SRR3099586_chr18
resources: tmpdir=/var/tmp/pbs.747714.pbsserver
[Wed Feb 21 13:37:54 2024]
Finished job 2.
2 of 8 steps (25%) done
Select jobs to execute...
Execute 1 jobs...
[Wed Feb 21 13:37:54 2024]
localrule fastqc:
input: Data/SRR3105697_chr18.fastq.gz
output: FastQC/SRR3105697_chr18_fastqc.zip, FastQC/SRR3105697_chr18_fastqc.html
log: Logs/SRR3105697_chr18_fastqc.std, Logs/SRR3105697_chr18_fastqc.err
jobid: 5
reason: Forced execution
wildcards: sample=SRR3105697_chr18
resources: tmpdir=/var/tmp/pbs.747714.pbsserver
[Wed Feb 21 13:38:02 2024]
Finished job 1.
3 of 8 steps (38%) done
Select jobs to execute...
Execute 1 jobs...
[Wed Feb 21 13:38:02 2024]
localrule fastqc:
input: Data/SRR3099587_chr18.fastq.gz
output: FastQC/SRR3099587_chr18_fastqc.zip, FastQC/SRR3099587_chr18_fastqc.html
log: Logs/SRR3099587_chr18_fastqc.std, Logs/SRR3099587_chr18_fastqc.err
jobid: 3
reason: Forced execution
wildcards: sample=SRR3099587_chr18
resources: tmpdir=/var/tmp/pbs.747714.pbsserver
[Wed Feb 21 13:38:04 2024]
Finished job 5.
4 of 8 steps (50%) done
Select jobs to execute...
Execute 1 jobs...
[Wed Feb 21 13:38:04 2024]
localrule fastqc:
input: Data/SRR3099585_chr18.fastq.gz
output: FastQC/SRR3099585_chr18_fastqc.zip, FastQC/SRR3099585_chr18_fastqc.html
log: Logs/SRR3099585_chr18_fastqc.std, Logs/SRR3099585_chr18_fastqc.err
jobid: 4
reason: Forced execution
wildcards: sample=SRR3099585_chr18
resources: tmpdir=/var/tmp/pbs.747714.pbsserver
[Wed Feb 21 13:38:14 2024]
Finished job 3.
5 of 8 steps (62%) done
[Wed Feb 21 13:38:15 2024]
Finished job 4.
6 of 8 steps (75%) done
Select jobs to execute...
Execute 1 jobs...
[Wed Feb 21 13:38:15 2024]
localrule multiqc:
input: FastQC/SRR3099586_chr18_fastqc.zip, FastQC/SRR3105698_chr18_fastqc.zip, FastQC/SRR3099587_chr18_fastqc.zip, FastQC/SRR3099585_chr18_fastqc.zip, FastQC/SRR3105697_chr18_fastqc.zip, FastQC/SRR3105
699_chr18_fastqc.zip
output: multiqc_report.html, multiqc_data
log: Logs/multiqc.std, Logs/multiqc.err
jobid: 7
reason: Input files updated by another job: FastQC/SRR3105698_chr18_fastqc.zip, FastQC/SRR3099587_chr18_fastqc.zip, FastQC/SRR3099585_chr18_fastqc.zip, FastQC/SRR3105697_chr18_fastqc.zip, FastQC/SRR310
5699_chr18_fastqc.zip, FastQC/SRR3099586_chr18_fastqc.zip
resources: tmpdir=/var/tmp/pbs.747714.pbsserver
[Wed Feb 21 13:38:37 2024]
Finished job 7.
7 of 8 steps (88%) done
Select jobs to execute...
Execute 1 jobs...
[Wed Feb 21 13:38:37 2024]
localrule all:
input: FastQC/SRR3099586_chr18_fastqc.html, FastQC/SRR3105698_chr18_fastqc.html, FastQC/SRR3099587_chr18_fastqc.html, FastQC/SRR3099585_chr18_fastqc.html, FastQC/SRR3105697_chr18_fastqc.html, FastQC/SR
R3105699_chr18_fastqc.html, multiqc_report.html
jobid: 0
reason: Input files updated by another job: FastQC/SRR3099587_chr18_fastqc.html, multiqc_report.html, FastQC/SRR3105697_chr18_fastqc.html, FastQC/SRR3105699_chr18_fastqc.html, FastQC/SRR3099585_chr18_f
astqc.html, FastQC/SRR3099586_chr18_fastqc.html, FastQC/SRR3105698_chr18_fastqc.html
resources: tmpdir=/var/tmp/pbs.747714.pbsserver
[Wed Feb 21 13:38:37 2024]
Finished job 0.
8 of 8 steps (100%) done
Complete log: .snakemake/log/2024-02-21T133728.474300.snakemake.log
Observe the output
You can now see in the log that Snakemake registered the 2 processors:
Provided cores: 2. It’s difficult to see if your jobs are actually running simultaniously just by looking at the log but you can clearly see that Snakemake runs the first 2 jobs at the same time: Execute 2 jobs...Running Snakemake on 2 processors doesn’t reduce the computation time considerably in this case since we only have 6 input files to process and the tools that we are running are already quite fast to execute.
A little recap before moving on...
Local execution:
Everything we’ve seen up until now could technically also be run on a local computer, even what we’ve just seen above if you have several processors on your PC (and provided you have the necessary software installed).
There are better ways than
--cores to parallelise:We’ve also seen that using
--cores N (N being the number of processors to use), enables us to distribute the workload and, thus, to accelerate the computation. However, when running on a cluster, this is not the most optimal solution for parallelising your pipeline.Why? Let’s take what we just did as an example:
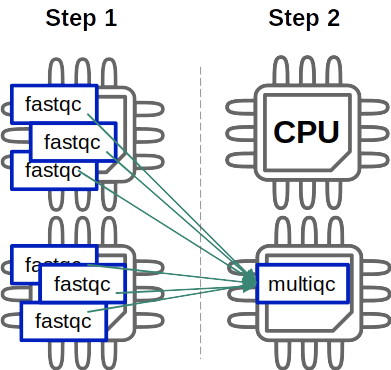
All 6 fastqc jobs can be evenly distributed over the 2 processors but the second rule (multiqc) is only using 1 of the 2 that we’ve reserved because there’s only one job for this rule. It’s not that bad when you’re using tools that run fast but becomes more problematic for software with much larger execution times…
What’s the solution? It would be more efficient to let the scheduler (OpenPBS in our case) deal with the distribution of jobs according to the resources available on the cluster. With this system, we’ll also be able to adapt the resources that are reserved to the amount each tool actually needs to run, thereby leaving unused resources free for others to use.