Getting started with Snakemake
Objective 2
Let’s scale up a little bit! Create a new snakefile named
ex1_o2.smk which will run FastQC on two RNA-seq input files.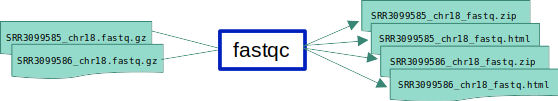
Where to start?
- Input files:
SRR3099585_chr18.fastq.gzandSRR3099586_chr18.fastq.gzin the${PWD}/Datadirectory - Expected output: we now expect result files for both of these inputs, so don’t forget to add the respective
*_fastqc.zipand*_fastqc.htmlfiles for the second input
Your Code for ex1_o2.smk should look like this:
rule fastqc: input:
"Data/SRR3099585_chr18.fastq.gz",
"Data/SRR3099586_chr18.fastq.gz"
output: "FastQC/SRR3099585_chr18_fastqc.zip", "FastQC/SRR3099585_chr18_fastqc.html",
"FastQC/SRR3099586_chr18_fastqc.zip",
"FastQC/SRR3099586_chr18_fastqc.html" shell: "fastqc --outdir FastQC/ {input}"
Test the script
Next, let’s check again if your pipeline still works:
snakemake -s ex1_o2.smk --cores 1 -p
-s : short form of the --snakefile option
-p: prints the commands
You should see something similar to the following output on your screen:
Assuming unrestricted shared filesystem usage for local execution.
Building DAG of jobs...
Using shell: /usr/bin/bash
Provided cores: 1 (use --cores to define parallelism)
Rules claiming more threads will be scaled down.
Job stats:
job count
------ -------
fastqc 1
total 1
Select jobs to execute...
Execute 1 jobs...
[Tue Feb 20 14:04:49 2024]
localrule fastqc:
input: Data/SRR3099585_chr18.fastq.gz, Data/SRR3099586_chr18.fastq.gz
output: FastQC/SRR3099585_chr18_fastqc.zip, FastQC/SRR3099585_chr18_fastqc.html, FastQC/SRR3099586_chr18_fastqc.zip, FastQC/SRR3099586_chr18_fastqc.html
jobid: 0
reason: Missing output files: FastQC/SRR3099586_chr18_fastqc.zip, FastQC/SRR3099586_chr18_fastqc.html
resources: tmpdir=/var/tmp/pbs.743371.pbsserver
fastqc --outdir FastQC/ Data/SRR3099585_chr18.fastq.gz Data/SRR3099586_chr18.fastq.gz
Started analysis of SRR3099585_chr18.fastq.gz
Approx 5% complete for SRR3099585_chr18.fastq.gz
Approx 10% complete for SRR3099585_chr18.fastq.gz
Approx 15% complete for SRR3099585_chr18.fastq.gz
Approx 20% complete for SRR3099585_chr18.fastq.gz
Approx 25% complete for SRR3099585_chr18.fastq.gz
Approx 30% complete for SRR3099585_chr18.fastq.gz
Approx 35% complete for SRR3099585_chr18.fastq.gz
Approx 40% complete for SRR3099585_chr18.fastq.gz
Approx 45% complete for SRR3099585_chr18.fastq.gz
Approx 50% complete for SRR3099585_chr18.fastq.gz
Approx 55% complete for SRR3099585_chr18.fastq.gz
Approx 60% complete for SRR3099585_chr18.fastq.gz
Approx 65% complete for SRR3099585_chr18.fastq.gz
Approx 70% complete for SRR3099585_chr18.fastq.gz
Approx 75% complete for SRR3099585_chr18.fastq.gz
Approx 80% complete for SRR3099585_chr18.fastq.gz
Approx 85% complete for SRR3099585_chr18.fastq.gz
Approx 90% complete for SRR3099585_chr18.fastq.gz
Approx 95% complete for SRR3099585_chr18.fastq.gz
Analysis complete for SRR3099585_chr18.fastq.gz
Started analysis of SRR3099586_chr18.fastq.gz
Approx 5% complete for SRR3099586_chr18.fastq.gz
Approx 10% complete for SRR3099586_chr18.fastq.gz
Approx 15% complete for SRR3099586_chr18.fastq.gz
Approx 20% complete for SRR3099586_chr18.fastq.gz
Approx 25% complete for SRR3099586_chr18.fastq.gz
Approx 30% complete for SRR3099586_chr18.fastq.gz
Approx 35% complete for SRR3099586_chr18.fastq.gz
Approx 40% complete for SRR3099586_chr18.fastq.gz
Approx 45% complete for SRR3099586_chr18.fastq.gz
Approx 50% complete for SRR3099586_chr18.fastq.gz
Approx 55% complete for SRR3099586_chr18.fastq.gz
Approx 60% complete for SRR3099586_chr18.fastq.gz
Approx 65% complete for SRR3099586_chr18.fastq.gz
Approx 70% complete for SRR3099586_chr18.fastq.gz
Approx 75% complete for SRR3099586_chr18.fastq.gz
Approx 80% complete for SRR3099586_chr18.fastq.gz
Approx 85% complete for SRR3099586_chr18.fastq.gz
Approx 90% complete for SRR3099586_chr18.fastq.gz
Approx 95% complete for SRR3099586_chr18.fastq.gz
Analysis complete for SRR3099586_chr18.fastq.gz
[Tue Feb 20 14:05:02 2024]
Finished job 0.
1 of 1 steps (100%) done
Complete log: .snakemake/log/2024-02-20T140449.328558.snakemake.log
Observe the output
As you can see in the highlighted line above, Snakemake detects that FastQC wasn’t yet run on your second input (“Missing output files: FastQC/SRR3099586_ch18_fastqc.html, FastQC/SRR3099586_ch18_fastqc.zip“, line 20) and then re-executes the fastqc command on this input.
Have a look at your output folder, you should now have 4 files in there:
john.doe@node06:/data/work/I2BC/john.doe/snakemake_tutorial$ ls FastQC
SRR3099585_chr18_fastqc.html SRR3099585_chr18_fastqc.zip SRR3099586_chr18_fastqc.html SRR3099586_chr18_fastqc.zip