The objective of this exercise is to adapt an already-existing bash pipeline into a Snakemake one
Our final objective is to create a pipeline with the following general steps:
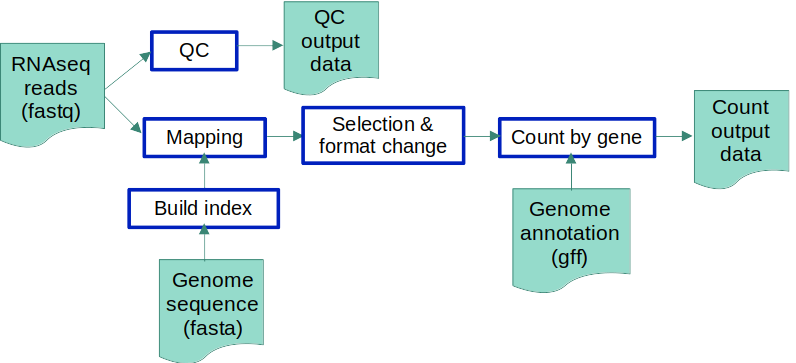
As you can see, not all pipelines are linear… Here, there are clearly 2 “independent” branches: the one that generates the final QC data of the RNAseq input files and the one that creates feature count tables using the RNAseq data as well as reference genomes and their annotation.