Learning how to manipulate and visualise protein structures
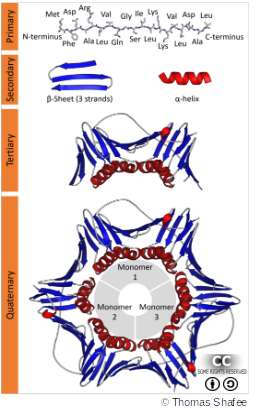
Proteins - from 1D to 4D
Proteins are made of chains of amino acids with different properties (hydrophobic, acidic, basic, …). These properties determine if and how they fold into 3D space. In this, different forces play a role:
- Primary structure: peptide (covalent) bonds between amino acids
- Secondary structure: hydrogen bonds between within amino acid backbones
- Tertiary & Quaternary structures: hydrophobic interactions for protein folding, hydrogen & disulfide bonds for maintaining the stable structure
A summary of the main forces involved in protein folding & communication is detailed below, with ideas of distances between the chemical elements involved and their free energy.
| Interaction | Distance dependence | Typical distance | Free energy (bond dissociation enthalpies for the covalent bonds) |
|---|---|---|---|
| Covalent bond | - | 1.5Å | 356kJ/mole (610kJ/mole for a C=C bond) |
| Hydrogen bond | Donor (here N), and acceptor (here O) atoms <3.5Å | 3.0Å | 2-6kJ/mole in water 12.5-21kJ/mole if either donor or acceptor is charged |
| Disulfide bond | - | 2.2Å | 167kJ/mole |
| Salt bridge | Donor (here N), and acceptor (here O) atoms <3.5Å | 2.8Å | 12.5-17kJ/mole may be as high as 30kJ/mole for fully or partially buried salt bridges, less if the salt bridge is external |
| Long-range electrostatic interaction | Depends on dielectric constant of medium. Screened by water. 1/r dependence | Variable | Depends on distance and environment. Can be very strong in nonpolar region but very weak in water |
| Van der Waals interaction | Short range. Falls off rapidly beyond 4Å separation. 1/r^6 dependence | 3.5Å | 4kJ/mole (4-17 in protein interior) depending on the size of the group (for comparison, the average thermal energy of molecules at room temperature is 2.5 kJ/mole) |
Where to find protein structures?
Protein structures can be determined experimentally using:
- Crystallography (compute electronic density from Xray diffraction)
- Nuclear Magnetic Resonance (NMR, compute models from distance between amino acids)
- Cryogenic Electron Microscopy (cryoEM, compute electronic density from electronic microscopy pictures)
Or in silico with prediction methods (de novo or based on existing 3D structures).
Several online databases of structures and models already exist & can be searched:
More about structural biology
To learn more or to address shortcomings in structural biology, please visit the Biostin-E website. It’s a free & open to all, do-it-yourself course. It judges your level through quizzes and redirects you to the videos you need in order to complete the knowledge that you’re missing in structural biology.
This free 3-hour online course provides an understanding of the fundamental concepts behind AlphaFold2, how users can run protein predictions and how AlphaFold2 has been used to enhance research.
“How to interpret AlphaFold structures” webinar from the European Bioinformatics Institute – EMBL-EBI.
The above course material is under CC-BY-SA license.