Learning how to manipulate and visualise protein structures
ChimeraX - the basics
ChimeraX is a programme for the interactive visualisation and analysis of molecular structures and related data, including density maps, trajectories, and sequence alignments. ChimeraX has a graphical interface and toolbar but is more easily used by typing commands in the command prompt directly as more functions and options are available that way.
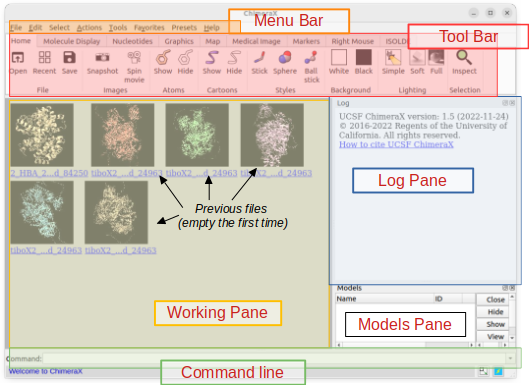
ChimeraX window layout
– log pane: The commands that you type or execute graphically through the menu bar will show up in the log pane and are clickable to open their respective help page.
– models pane: Molecules that are loaded into ChimeraX are listed in the models pane and are given an ID.
– working pane: The working pane is where you’ll be able to visualise your models. By default, you’ll see a list of models you’ve previously worked on (also accessible through the Tool bar in Home > Recent).
Other panes will appear/disappear according to your actions (e.g. in the Menu bar: Tools > Sequence > Show Sequence Viewer will open a new pane with the amino acid sequence of your models). They can be detached and/or moved around as you like.
Where to get ChimeraX help & tutorials
General help: If you have a look under Help in the Menu bar, you will find a link towards ChimeraX’s User Guide, Quick Start Guide and Tutorials (also accessible via the web). Once you’ve opened one of these dialog boxes, you can easily navigate through the docs using the integrated search bar (top left).
Help per function: Through the command line, you can access the help of each command by typing help your_cmd. You can also open the help page by clicking on the corresponding command in your command history in the log pane.
The above course material is under CC-BY-SA license.