Learning how to manipulate and visualise protein structures
Demo Step 2: Load the data in ChimeraX
Icon legend
![]() : the hand icon specifies graphical ways of running tools in ChimeraX that go through the Tool & Menu bars
: the hand icon specifies graphical ways of running tools in ChimeraX that go through the Tool & Menu bars
![]() : the terminal icon specifies in-line ways of running tools in ChimeraX that go through the command line pane at the bottom of the window
: the terminal icon specifies in-line ways of running tools in ChimeraX that go through the command line pane at the bottom of the window
Start ChimeraX and load your files
You can upload a structure file that is on your computer:
![]() graphically through the Tool bar in Home > Open or through the Menu bar File > Open (drag & drop should also work)
graphically through the Tool bar in Home > Open or through the Menu bar File > Open (drag & drop should also work)
![]() in-line with the open path/to/your/file command
in-line with the open path/to/your/file command
Of note: you can fetch structures directly from online databases in-line (if you have internet connection) with:
![]() open https://url/to/your/file
open https://url/to/your/file
e.g. open https://files.rcsb.org/download/8EF0.pdb to download the structure of rhMZ104-D antibody with PDB code 8EF0
![]() open dbname:proteincode (see help for more options)
open dbname:proteincode (see help for more options)
e.g. open alphafold:O15552 to load the Free fatty acid receptor 2 protein with UniProt code O15552 from the AlphaFold database
What you should now see
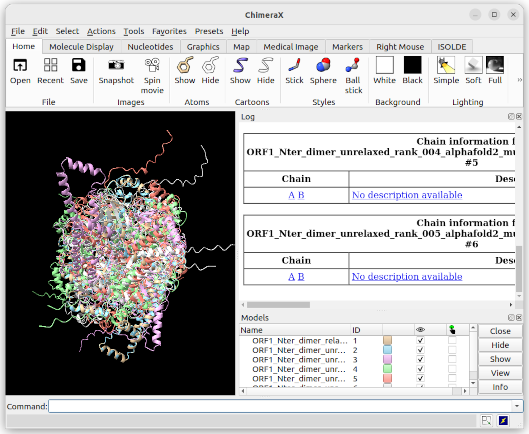
After uploading all 5 unrelaxed models and the relaxed model for rank 1, you should see:
- All 6 3D structures are shown in the Working pane
- These structures are also listed in the Models pane, and each of them is given a unique ID number
- The Log Pane shows you some more details about the structures you’ve uploaded (they all have 2 chains: A & B, for example), as well as the command you effectively used to upload your files – this is a good way to get familiar with the command line
You can show/hide elements by checking/unchecking the tick boxes in the Models pane (“eye” column) and select/deselect elements in the same way using the next column on the right. You can show/hide all by using the buttons on the far right of the Models pane. Similarly, you can delete elements using the Close button.
Note: the Models pane will also harbour the list of other elements you will generate (e.g. interaction elements, distance elements, etc.). They will also all be attributed unique ID numbers.
The above course material is under CC-BY-SA license.