Learning how to manipulate and visualise protein structures
Demo Step 3: Getting familiar with ChimeraX
Below is a list of useful commands to manipulate your structure in ChimeraX. Try playing around with these functions (practice makes perfect!).
Mouse commands to manipulate structures
![]() Move your structures around in the Working pane:
Move your structures around in the Working pane:
![]() Left click + hold on the protein(s) to move it about on vertical & horizontal axis
Left click + hold on the protein(s) to move it about on vertical & horizontal axis
![]() Left click + hold on the background to rotate it about clockwise or anticlockwise along a fixed axis
Left click + hold on the background to rotate it about clockwise or anticlockwise along a fixed axis
![]() Left click + Shift + hold to translate the protein
Left click + Shift + hold to translate the protein
![]() Scroll to zoom in & out
Scroll to zoom in & out
![]() Select & deselect regions of your structure in the Working pane:
Select & deselect regions of your structure in the Working pane:
![]() Left click + Ctrl + drag over an area of your protein to select this region
Left click + Ctrl + drag over an area of your protein to select this region
![]() Left click + Crtl + click a specific residue to select a specific residue
Left click + Crtl + click a specific residue to select a specific residue
![]() Left click + Ctrl + Shift + an area/residue to add to or remove from the current selection depending if the area/residue is already in the selection or not
Left click + Ctrl + Shift + an area/residue to add to or remove from the current selection depending if the area/residue is already in the selection or not
![]() Left click + Crtl + click on the background to clear the current selection
Left click + Crtl + click on the background to clear the current selection
![]() Use Up/Down arrow keys to expand the current selection to the lower/higher resolution level (e.g. from a residue to the whole secondary structure it is contained in)
Use Up/Down arrow keys to expand the current selection to the lower/higher resolution level (e.g. from a residue to the whole secondary structure it is contained in)
Note: to select at the atomic level (with your mouse), you will need to change the resolution of the protein representation first.
![]() You can also define the function of your Right click through the Tool bar under the Right Mouse tab:
You can also define the function of your Right click through the Tool bar under the Right Mouse tab:
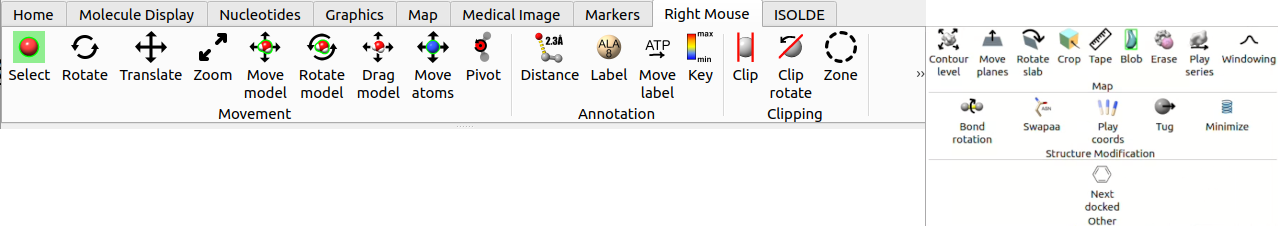
By default, the Right click is in Translate mode (green background = selected) but you can also change it to Zoom for example if you don’t have a Scroll button handy.
![]() Click here for detailed information on the features in the Right Mouse tab.
Click here for detailed information on the features in the Right Mouse tab.
![]() In-line notation conventions – specifying models, chains and residues
In-line notation conventions – specifying models, chains and residues
There are conventions to specify or target structures and/or regions of interest in your commands. Different level notations are bricks that can be combined together.
For example: # followed by the model ID will specify the model. # alone selects all models loaded in ChimeraX. To specify several models, use the comma as separator (OR operator) e.g. #1,5 will select models 1 and 5. To select elements respecting 2 different conditions, use & (AND operator).
| Levels | Models | Chains | Residues | Atoms |
|---|---|---|---|---|
| Symbol | # | / | : | @ |
| Usage examples | #1 = model 1 #1,5 = model 1 & 5 |
/A = all chains A #1/A,B = chains A & B of model 1 only |
:1-25 = all residues with numbers 1 to 25 :tyr = all Tyrosines #1/A:1-25&:tyr = all Tyrosine residues with numbers between 1 and 25 and who belong to chain A of model 1 |
@ca = all C alpha atoms #1@ca = all C alpha atoms of model 1 |
The above course material is under CC-BY-SA license.